Biochemical Tests
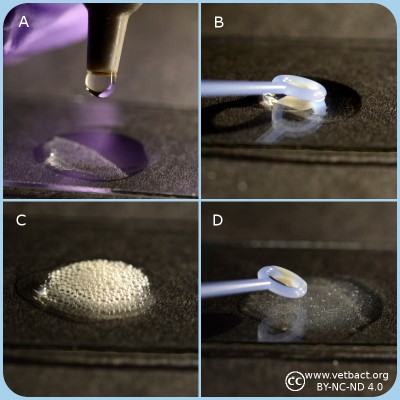
Catalase test
Catalase test
A. Apply one drop of 3% hydrogene peroxide on a microcscopic slide.
B. Transfer bacteria with a plastic loop to the H2O2 solution.
C. Catalase positive bacteria produce gas (O2) in form av bubbles which shows that the bacterium has a catalase.
D. Catalase negative bakteria does not have a catalase and therefore oxygene is not formed. Note the fragments of bacterial colonies, but there are no bubbles in the solutions.
- Click on the image to enlarge it.
General information
Many aerobic bacteria and most of those which are facultatively anaerobic produce the enzyme catalase. The function of this enzyme is to detoxify hydrogenperoxide (H2O2), which is formed from the superoxide radical by superoxide dismutase. Many aerotolerant anaerobic bacteria have peroxidase (which is not the same enzyme as cytochrome c oxidase) instead of catalase. Obligate anaerobic bacteria lack superoxide dismutase and catalase.
Catalase contains a heme group at the active site and it is catalyzing the following reaction with a very high turnover number:
2 H2O2 → 2 H2O + O2
Method
- Spread the bacteria on an agar plate and inkubate the plate over night (18-24 h) under appropriate conditions.
- Collect bacteria from one colony with a sterile inoculating loop (of plastic or platinum) and apply the bacteria on a microscope slide.
- If the bacteria are collected from a blood agar plate, one has to avoid contamination of agar, because hemoglobin also contains heme groups which can cause a false positive reaction.
- Add one drop of 3% H2O2 to the bacteria and observe the suspension. Be careful with the handling of H2O2 which is corrosive!!!
- Positive test resultat: Gas formation (O2) in the form av bubbles shows that the bacterium has a catalase.
- Negative test resultat: No gas formation.
Applications
The catalase test is primarily used for gram positive bacteria and can for instance be utilized to distinguish Staphylococcus spp. and Micrococcus spp., which are catalase positive from Streptococcus spp. and Enterococcus spp., respectively, which are catalase negative.
Updated: 2017-12-20.
ContentsCitrate test
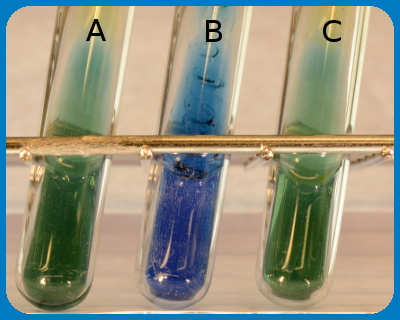
The figure shows the result of a citrate test , by deep inoculation into "Simmons citrate tubes", where tube A contains a negativ control (no bacteria). Tube B contains Citrobacter freundii and tube C contains Escherichia coli. Note that a colour change has occured in tube B because C. freundii can grow on citrate as carbon source.
Image: Karl-Erik Johansson (BVF, SLU) and Lise-Lotte Fernström (BVF, SLU).
General
Some bacteria can utilize citrate as the only carbon source and the citrate test shows if the actual bacterium has this capability.
Method
- Inoculate a tube containing citrate medium with a small amount of bacteria. It is also possible to streak or perform a deep inoculation into "Simmons citrate tube".
- Incubate at 30-37ºC during 24-48 h.
- Positive test result: growth in citrate medium or growth with colour change to blue in Simmon's citrate tube.
- Negative test result: no growth in citrate medium eller growth but no colour change (still green colour) in Simmon's citrate tube.
Use
The citrate test is used to distinguish between, among others Citrobacter freundii and Escherichia coli.
Updated: 2018-06-06.
ContentsCoagulase test
General
Some bacteria produce coagulase, which is an enzyme that converts fibrinogen to fibrin, which means that it can coagulate plasma. The ability to produce coagulase is assumed to be associated to the virulence of staphylococci. The test is used to distinguish between coagulase positive and coagulase negative staphylococci.Method
- Suspend one colony from the suspected pure culture in 0.5 ml of plasma from horse, rabbit or man.
- Incubate at 37ºC.
- Read the test after 4 h. If the result is negative (see below), continue with the incubation.
- Perform the final read after 24 h.
- Positive reaction if the plasma coagulates and the coagulate is stable. It must not be dissolved upon stirring.
- Negative reaction if the plasma does not coagulate or if the coagulate is dissolved again upon stirring.
Coagulase test of Stahylococcus spp. The upper tube shows positive result (the plasma has coagulated) and the lower tube shows a negative result.
Use
The coagulase test is used to distinguish between Staphylococcus aureus from coagulase negative Staphylococcus spp. Note, however, that some strains of S. aureus can be coagulase negative, but it is unusual. Some strains of S. hyicus and S. intermedius can br coagulase positve. S. pseudintermedius is coagulase positive, but not until after 24 h.
Updated: 2013-10-01.
ContentsDNase test
General
Many bacteria have enzymes that break down nucleic acids. The bacteria can then use the resulting nucleotides to build up their own nucleic acids. DNase is such an enzyme, which thus hydrolyzes DNA. Existence of DNase is characteristic for certain species or strains of bacteria and can be used for typing.
Method
Figure: DNase test of Staphylococcus spp. The strain in the upper streak is negative (no clearing around the streak), whereas the strain in the lower streak is positive. (Image: SLU/SVA)
Presence of DNase can be determined by cultivation on an agar plate, which contains DNA. If the bacterium has DNase and if the bacteria are allowed to grow over night, the DNA will be hydrolyzed into the constituting nucleotides. Diluted hydrochloric acid (HCl) is then poured onto the plate and there will be a clear zone close to the colonies or the streak, because individual nucleotides are soluble in diluted HCl, but not DNA, which precipitates in the rest of the plate.
Use
The test is useful to distinguish between:
- Serratia spp. and Enterobacter spp.
- Staphylococcus aureus (most strains are coagulase positive) and coagulase negative Staphylococcus spp.
- Moraxella catarrhalis and Neisseria spp.
Updated: 2017-12-08.
ContentsHippurate test

The figure shows the result of an hippurate test , where tube A is a negative control. Tube B contains Streptococcus agalactiae, which is hippurate positive and tube C contains Streptococcus dysgalactiae subsp. dysgalactiae, which is hippurate negative. Note that a colour change has occured in tube B because S. agalactiae is hippurate positive. Observe also that hippurate negative bacteria may give a faint colour change.
Image: Karl-Erik Johansson (BVF, SLU) och Lise-Lotte Fernström (BVF, SLU).
General
Some bacteria can hydrolyze hippurate to the amino acid glycine and benzoate by means of the enzyme hippuricase. Glycine can be detected with ninhydrin (2,2-Dihydroxyindane-1,3-dione), which reacts with free amino groups (-NH2) and a blue product is formed.
Method
- Suspend a loop-full of bacteria (for instance Campylobacter sp.) in 0.5 ml of sodium hippurate siolution.
- Incubate the suspensionen at 37ºC during 2 h in a water bath.
- Then carefully add 0.2 ml of ninhydrin solution without mixing.
- Incubete the dube during another 10 min. at 37ºC before reading the result.
- Positive test resultat: Deep blue colour.
- Negative test result: Pale blue colour.
Use
The hippurate test is primarely used to distinguish between Campylobacter jejuni (hip+) and Campylobacter coli (hip-) and to distinguish between different streptococci (see figure).The test is also used, in combination with other methods, to type Brachyspira spp.
Updated: 2017-03-31.
ContentsHydrogen sulfide production
General
Some bacteria can metabilize certain sulfur containing compounds under production of hydrogen sulfide (H2S). Hydrogen sulfide is a toxic, flamable and badly smelling gas (smells like rotten eggs). If soluble iron or lead salts (for instance ferric citrate) is used in a so-called H2S-medium, which should also contain sodium thiosulfate (Na2S2O3), they can react with H2S, if present, under formation of black insoluble iron and lead sulfide, respectively.Method
- Perform a deep inoculation in the H2S medium with bacteria from one colony.
- Incubate the tube at 30-37ºC during 24-48 h.
- Positive test result: a black precipitate in the medium.
- Negative
test result: no precipitate in the medium.
Use
The test can be used for differentiation of, among other bacteria, certain Campylobacter spp.
Updated: 2012-12-11.
ContentsIndole test
General
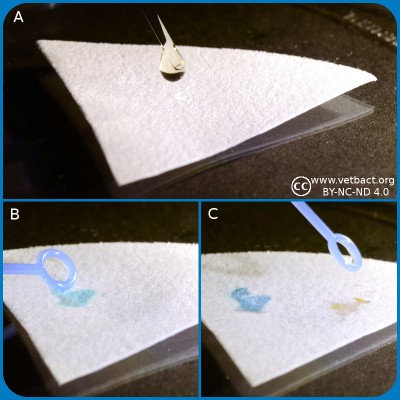
Spot indole test.
A. Application of spot indole reagent.
B. Application of indole positive bacterium.
C. Application of indole negative bacterium.
Click on the image to enlarge it.
Bacteria, which express the enzyme tryptophanase can hydrolyze the amino acid tryptophan to indole, pyruvic acid and ammonia. Presence of indole can be shown by means of Kovác's reagent or by spot indole test. In the spot test indole reacts with p-Dimethylaminocinnamaldehyde to produce a blue to blue-green product. Kovác's reagent contains p-dimethylaminobenzaldehyde, which forms a red complex with indole.
Method, spot indole test
- Place several drops of Spot Indole Reagent on a piece of filter paper.
- Fill a plastic loop with bacteria from a colony cultivated for 24-48 h.
- Rub it onto the reagent saturated area of the filter paper.
- Positive reaction: Appearance of a blue to blue-green color change within 10 seconds.
- Negative reaction: Remain colorless or light pink.
Method, Kovac´s reagent
- Suspend one colony from a pure culture of the bacterium to be investigated, in a suitable medium (for instance LTLSB or tryptophan medium).
- Incubate the medium at 37°C during 20-28 h.
- Add a few drops of Kovác's reagent.
- Positive test result: The indole reagent change colour to cerise red.
- Negative test result: The indole reagent remains pale yellow.
Use
Confirmation of suspected E. coli-strains. Typing (species determination) of Brachyspira spp. in combination with other tests. Kovac's indole reagent is more sensitive than the indole spot reagent, but it is not recommended for use with anaerobic bacteria. The indole spot reagen is suitable for both aerobic and anaerobe use.
Updated: 2017-12-20.
ContentsLecithinase test
General
Many bacteria have enzymes which can break down lipids, so-called lipases. Lecithinase, which is also called phospholipase C, is such an enzyme that splits the phospholipid lecithin (= e.g. phosphatidylcholine). Phospholipids, which are charged are usually soluble in water, but one of the products which is formed by the splitting, namely a diglyceride, is not charged and it has two long hydrocarbon chains. It is, therefore, unsoluble in water and this is utilized in the lecithinase test, where bacteria are cultivated on egg yolk agar. Egg yolk contains a lot of lecithin.Method
- Apply the bacteria in the form of a streak onto the egg yolk agar.
- Read the plate after 24 h.
- Positive test result: Precipitation around the streak of bacteria.
- Negative test result: No precipitation.
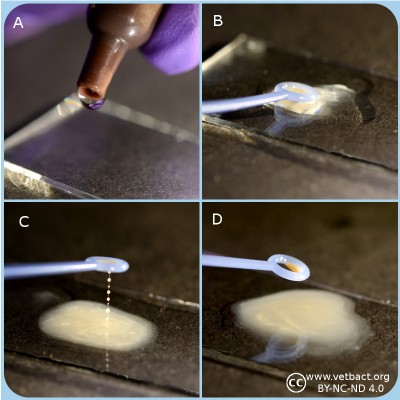
Figure: Bacillus cereus on egg yolk agar. Note the precipitation around the streak of bacteria, which shows that they are positive for lecithinase.
Use
Can among other things be used to differentiate between certain species within the genus Bacillus.
Updated: 2012-12-11.
ContentsMixed acid fermentation
General
Some bacteria can ferment glucose to a mixture of the following organic acids: formic acid, acetic acid and lactic acid. This is called mixed acid fermentation and it causes highly decreased pH in the medium. Mixed acid fermentation can, therefore, be detected by addition of the pH indicator methyl red (MR). The test method is sometimes called the MR test.
Method
Methyl red test, where a positive reaction has occured in the right tube. (Image: SLU/SVA.)
- Suspend a bacterial colony from a pure culture in the MR/VP medium.
- Incubate at 30°C during 3 days or at 37°C during 48 h.
- Add 2-3 drops of a solution of methyl red.
- Positive test result: red colour change
- Negative test result: no colour change.
Use
Some members of the family Enterobacteriaceae have mixed acid fermentation (see the respective bacterial page), which can be used to differentiate these bacteria.
Updated: 2017-12-08.
ContentsOxidase test
Oxidase test
A. Two drops of oxidase reagent is applied onto a piece of filter paper.
B and C. Colony material is transferred by a plastic loop to the spot of oxidase on the filter paper.
C. The left spot includes oxidase positive bacteria and the spot to the right oxidase negative bacteria.
D. The final results can be observed after 30 s.
- Click on the image to enlarge it.
General information
Bacteria, which have aerobic respiration, often have cytochrome c and a cytochrome c oxidase. The presence of these components can in combination with other methods be used for typing. A commersial test, which contains an artificial electron acceptor (N, N, N', N'-tetramethyl-p-phenylenediamine, see Fig. 1), is often used. This artificial electron acceptor change colour depending upon redox state. The substance is also referred to as a redox indicator and it can be oxidized by the oxidized form of cytochrome c. Cytochrome c oxidase is the last enzyme of the electron transport chain, where it normally reduces oxygen to water and pump protons to the outside according to the following net reaction:
4 Fe2+-cytochrome cred + 8H+in + O2 → 4 Fe3+-cytochrome cox + 2H2O + 4H+out
Cytochrome c oxidase is a transmembrane protein complex (Complex IV), which is also present in the cytoplasmic (inner) membrane of mitochondria.
Method
- Keep the plastic ampoule (which contains a glass ampoule) between your fingers and with the opening upwards and away from your face. Press the ampouls with your fingers until the glass ampoule breaks.
- Apply two drops of the oxidase reagent onto a piece of filter paper.
- Transfer bacteria from one colony with a plastic or platinum loop onto the spot with the oxidase reagent. The colonies should have been incubated at the appropriate temperature for 18-24 h .
- Positive test resultat: Dark blue-purple colour change within 10-30 sec.
- Negative test resultat: No colour change or colour change after more than 30 sec.
Note that the oxidase reagent is not stable after that the ampoule has been opened. It may be used for a couple of hours, but eventually it will be oxidized by the oxygen in the air.
Applications
The oxidase test is used for identification of gram negative bacteria. For instance to identify members of the family Enterobacteriaceae, which are oxidase negative, except members of the genus Plesiomonas (oxidase positive). Members of the family Pseudomonadaceae, and the genera Aeromonas and Campylobacter are oxidase positive.
Updated: 2017-12-20.
ContentsPotassium hydroxide test
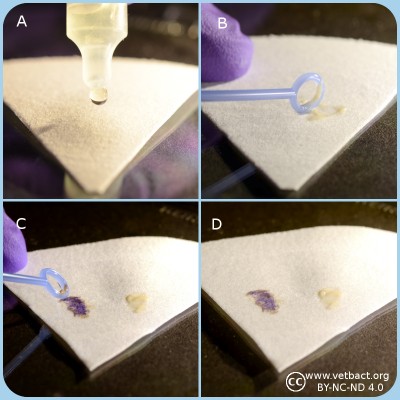
Potassium hydroxide test.
A. Application of potassium hydroxide solution onto a microscopic slide.
B. Bacteria are transferred and mixed with the KOH solution.
C. Result with gramnegative bacteria where the solution will be viscous and form a mucoid string.
D. Result with grampositive bacteria where the solution will not be viscous.
Click on the image to enlarge it.
General
The purpose of the potassium hydroxide test (KOH test) is to identify gram negative bacteria. KOH dissolves the thin layer of peptidoglycan of the cell walls of gram negative bacteria, but does not affect gram positive cell walls. Disintergration of gram negative cell walls lyses the cell and release its contents, including the DNA. The DNA will make the solution very viscous and the solution will stick to the plastic loop when touched. Gram positive bacteria will not be affected by KOH, because they have thicker peptidoglycan layer in the cell wall. Thus, the cells will not be lysed, the DNA not released and no viscosity will be observed.
Material
- Microscopic slide
- Plastic loop
- 3% KOH
Method
- Apply one drop of 3% KOH on a microscopic slide.
- Use o loop to transfer a generous amount of bacteria (cultivated for 24-48 h) to the drop of KOH
- Stir carefully
- The solution of gram negative bacteria will be viscous and form a mucoid string within 30 sec
- Use positive and negative controls
Positive results: The solution with the bacteria (gram negative) will be viscous
Negative results: The solution with the bacteria (gram positive) will not be viscous
Use
The purpose of the KOH test is to quickly distinguish between gram negative and gram positive bacteria as a complement to Gram staining. The test is not useful for anaerobic bacteria.
Updated: 2017-12-20.
ContentsUrease test
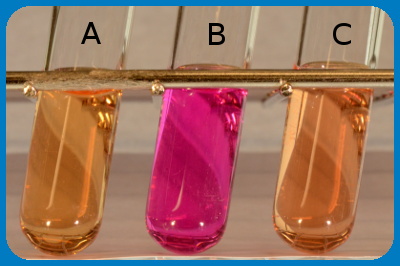
The figure shows the result of an urease test, where tube A is a negative control. Tube B contains, Proteus sp. and tube C contains Escherichia coli. Note that a pH change has occured in tube B because Proteus spp. are urease positive.
Image:Karl-Erik Johansson (BVF, SLU) and Lise-Lotte Fernström (BVF, SLU).
General
Some bacteria have the enzyme urease, which in the presence of H2O converts urea (=carbamide) to NH3 (ammonia) and CO2 (carbondioxide), which forms ammonium carbonate in the presence of water. See chemical reaction formula.
Method
By growing the bacteria in urease medium containing a pH indicator, it can be determined if the bacteria express urease. If the bacteria have an urease, urea will be converted to ammonium carbonate and the medium will turn alcaline. Thus, the colour will change to red (cerise).
Urease medium:
| Substance | Amount (g) |
|---|---|
| Pepton | 1.0 |
| Glucose | 1.0 |
| NaCl | 5.0 |
| Disodium hydrogen phosphate | 1.2 |
| Potassium dihydrogen phosphate | 0.8 |
| Phenol red | 0.004 |
Applications
The urease test can be used to distinguish between E. coli, which is urease negative, from Proteus spp., which are urease positive.
Updated: 2017-03-31.
ContentsVoges-Proskauer (VP) test
General
The VP test shows if the bacterium has butanediol fermentation and can split glucose to acetoin via pyruvat and further to 2,3-butanediol according to:2 pyruvate + NADH --> 2CO2 + 2,3-butanediol.
If KOH (potassium hydroxide) is added, acetoin will be converted to diacetyl (= 2,3-butanedione), which reacts with alpha-naphtol and forms a pink complex.Method
- Suspend one colony from the pure culture, which is to be investigated, in VP/MR medium.
- Incubate at 30-37ºC during 24-48 h.
- Add 0.2 ml of 40% KOH and then 0.6 ml of
alpha-naphtol solution.
- Positive test result: colour change to pink.
- Negative
test result: no colour change.
Voges-Proskauer test, where the tube to the right shows a positive test resultt. (Image: SLU/SVA.)
Use
Klebsiella spp. and Enterobacter spp. has the capacity to perform butanediole fermentation in contrast to Escherichia coli, Salmonella spp. and Shigella spp.
Updated: 2012-12-11.
Contents